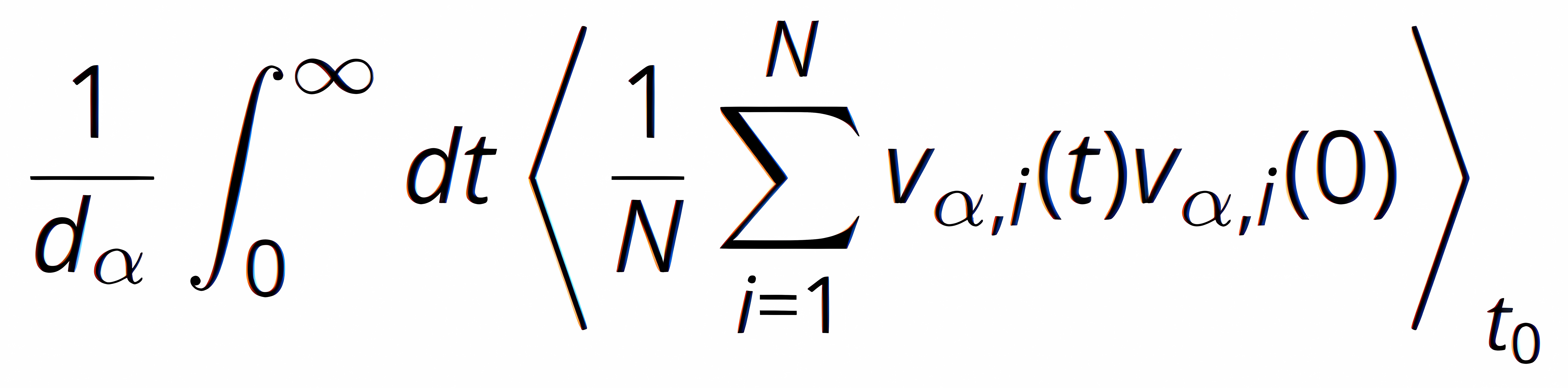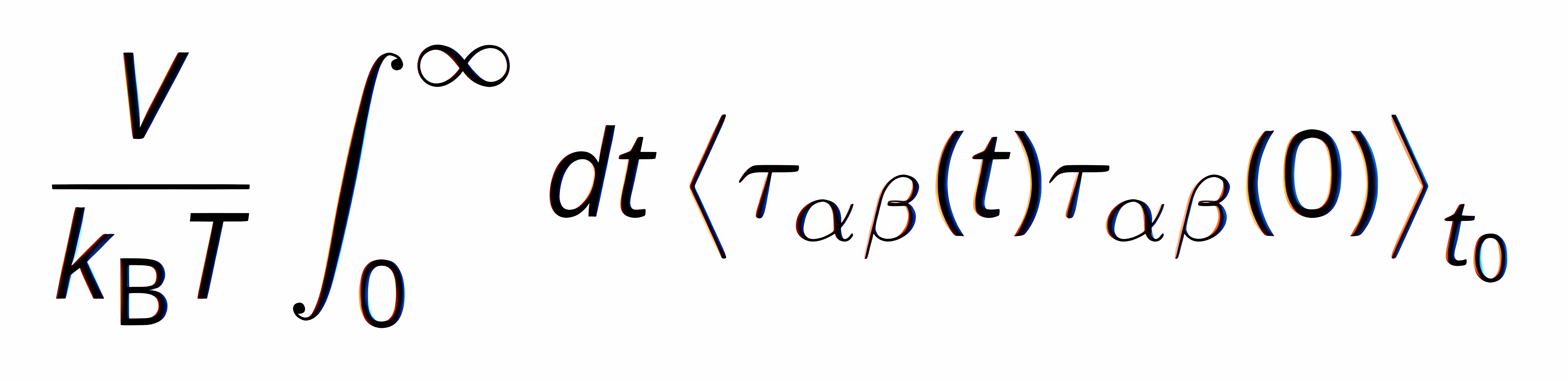Rationale
Transport properties are among the most requested computations in the MDAnalysis project. These measurable values defining the transfer of mass, momentum, heat, and charge are widely used in biomolecular research and chemical engineering.1, 2 Since molecular dynamics can predict transport properties with good accuracy and reliability,1 it would be extremely useful for users to have access to easy-to-use implementations of these complex calculations. My Google Summer of Code (GSoC) project aims to add methods for calculating self-diffusivity and viscosity, the two most common transport properties in the literature.1 I will take care in testing the calculations and writing accessible documentation to make the analyses approachable for everyone.
Project Deliverables
- Construct a class to calculate self-diffusivity via the Green-Kubo method
- Write a utility function to calculate shear viscosity via the Einstein method
- Create a utility function to calculate shear viscosity via the Green-Kubo method
- Test and document the new features thoroughly, ensuring the calculations are reliable and easy to use
Self-Diffusivity
As described by Maginn et al. (2018), self-diffusivity will be calculated via the Green-Kubo approach as follows:
where $N$ is the number of molecules, $d$ is the dimensionality, and $v$ denotes velocity.1 Figure taken from Best Practices for Computing Transport Properties.
The angle brackets contain a velocity autocorrelation function (VACF), an autocorrelation function (ACF) for velocity. In general, an ACF can be calculated with:
where $N$ is the number of time frames and $\Delta t$ are discrete time intervals between data points.3 Figure taken from the GROMACS Reference Manual.
In practice, the Green-Kubo integral reaches a plateau i.e. the VACF decays to 0. Such a time would then be the upper limit of integration, rather than $\infty$. Adapting the mean squared displacement (MSD) module from MDAnalysis would provide a reliable class to calculate the VACF. The tidynamics package will provide a Fast Fourier Transform (FFT) algorithm to compute mean squared displacement (MSD) with $O(nlogn)$.4,5
Alternatively, users can choose the simple “windowed” method with $O(n^2)$, as is possible in the MSD module.
Users will then call scipy.integrate.trapezoid() to compute the integral and divide by the dimensionality to obtain the self-diffusivity.6
Viscosity
Shear viscosity will be calculated via the Einstein method below:
where $V$ is volume, $k_B$ is the Boltzmann constant, $T$ is temperature, and $\tau _ {\alpha \beta}$ is a general term for components of the pressure tensor, with $\alpha \ne \beta$.1 Figure taken from Best Practices for Computing Transport Properties.
Shear viscosity will also be calculated using the Green-Kubo method:
with the same terms defined in the Einstein approach for shear viscosity.1 Figure taken from Best Practices for Computing Transport Properties.
Because MDAnalysis does not currently support pressure/stress tensors for most file types, a practical solution is to provide a utility function to calculate shear viscosity from a NumPy array of the pressure tensor. For GROMACS simulations, the EDRReader in MDAnalysis can obtain pressure tensor components, temperature, and volume directly from EDR files.7 For simulations from other MD packages, the user would input this information in the utility function.
Einstein Implementation Outline
We square the integral of the pressure tensor, then divide by the number of pressure tensor components chosen for averaging.1,8,9 We store these results in a NumPy array. We then multiply these results at different time lengths by the average volume, then divide that by the Boltzmann constant, the average temperature, and the corresponding times. This will give a NumPy array of “viscosity” over time, which is meant to give an idea of the viscosity rather than be used as a definitive result. The utility function will also return the corresponding times as a NumPy array so that the user can easily visually assess the results and determine where they feel is most appropriate to take the slope based on their data.1
Green-Kubo Implementation Outline
This implementation will be quite similar to the last two conceptually. It will also take the form of a utility function and handle the pressure tensor and inputs in a similar manner to the Einstein implementation for viscosity. The pressure autocorrelation function will be calculated as explained in the self-diffusivity outline but using pressure tensor components instead of velocities. Likewise, tidynamics.acf() will provide an FFT option here. Finally, we will use scipy.integrate.cumulative_trapezoid() to integrate the pressure autocorrelation function at increasing time lengths, providing the user a “viscosity” array corresponding to those time lengths that they would be able to evaluate.10
First Meeting and Next Steps
I had the pleasure of finally speaking face to face with @hmacdope and @orionarcher on May 11. Hugo and Orion continue to be extremely welcoming and supportive. They were among the MDAnalysis developers who reviewed my first ever open source contributions, and I have really enjoyed working with them. We decided that my project will likely work best as an MDAKit, a standalone package that uses the MDAnalysis library. They provided some valuable tips and action items for the next while, summarized here:
- Set up blog
- Attend MDAKits meeting, read the docs, and try the cookiecutter
- Discuss implementing an Einstein and Green-Kubo backend
- Look into using the Helfand method for viscosity
- Interact with the community (Discord, mailing lists, PRs)
I have made some progress on the blog, planning the backend, reading about MDAKits, and interacting with the community. However, there is much more to be done with fleshing out my plans and researching the Helfand method. It would be nice to have an MDAKit repository set up soon as well. The coding period is fast approaching, so look forward to another update in a few weeks.
References
Maginn, E. J.; Messerly, R. A.; Carlson, D. J.; Roe, D. R.; Elliot, J. R. Best Practices for Computing Transport Properties 1. Self-Diffusivity and Viscosity from Equilibrium Molecular Dynamics [Article v1.0]. Living J. Comput. Mol. Sci. 2018, 1 (1), 6324–6324. https://doi.org/10.33011/livecoms.1.1.6324. ↩ ↩2 ↩3 ↩4 ↩5 ↩6 ↩7 ↩8
Wang, J.; Hou, T. Application of Molecular Dynamics Simulations in Molecular Property Prediction II: Diffusion Coefficient. J. Comput. Chem. 2011, 32 (16), 3505–3519. https://doi.org/10.1002/jcc.21939. ↩
Correlation functions. GROMACS 2023 documentation. https://manual.gromacs.org/documentation/current/reference-manual/analysis/correlation-function.html (accessed 2023-03-21). ↩
Buyl, P. de. Correlation routines. tidynamics 1.0.1 documentation. https://lab.pdebuyl.be/tidynamics/correlation.html (accessed 2023-03-19). ↩
Buyl, P. de. Tidynamics: A Tiny Package to Compute the Dynamics of Stochastic and Molecular Simulations. J. Open Source Softw. 2018, 3 (28), 877. https://doi.org/10.21105/joss.00877. ↩
scipy.integrate.trapezoid. SciPy v1.10.1 Manual. https://docs.scipy.org/doc/scipy/reference/generated/scipy.integrate.trapezoid.html (accessed 2023-03-22). ↩
Auxiliary files: EDR Files. MDAnalysis User Guide documentation. https://userguide.mdanalysis.org/stable/formats/auxiliary.html#edr-files (accessed 2023-04-02). ↩
Gonzalez-Salgado, D.; Vega, C. A New Intermolecular Potential for Simulations of Methanol: The OPLS/2016 Model. J. Chem. Phys. 2016, 145 (3), 034508. https://doi.org/10.1063/1.4958320. ↩
Liu, H.; Maginn, E.; Visser, A. E.; Bridges, N. J.; Fox, E. B. Thermal and Transport Properties of Six Ionic Liquids: An Experimental and Molecular Dynamics Study. Ind. Eng. Chem. Res. 2012, 51 (21), 7242–7254. https://doi.org/10.1021/ie300222a. ↩
scipy.integrate.cumulative_trapezoid. SciPy v1.10.1 Manual. https://docs.scipy.org/doc/scipy/reference/generated/scipy.integrate.cumulative_trapezoid.html (accessed 2023-03-26). ↩